Help and Tutorial
What is NBDB database?
Binding of nucleotide-containing ligands is an indispensable element of many biochemical functions,
and evolution of protein-nucleic acids interactions started from the very origin of life.
Not surprisingly, analysis of protein-nucleotide interactions is a topic of numerous studies.
This database is the first comprehensive collection of conserved sequence profiles in form of Position-Specific Scoring Matrices (PSSMs) of protein motifs,
Elementary Functional Loops (EFLs), and their position-specific interactions with specific chemical moieties in 24 major nucleotide-containing ligands and biologically relevant cofactors with molecular weight less than 1000 g/mol (see Table below).
Users can search for EFL profile representatives within the sequences/structures of interest. The sitemap below shows NBDB pages and links between them.
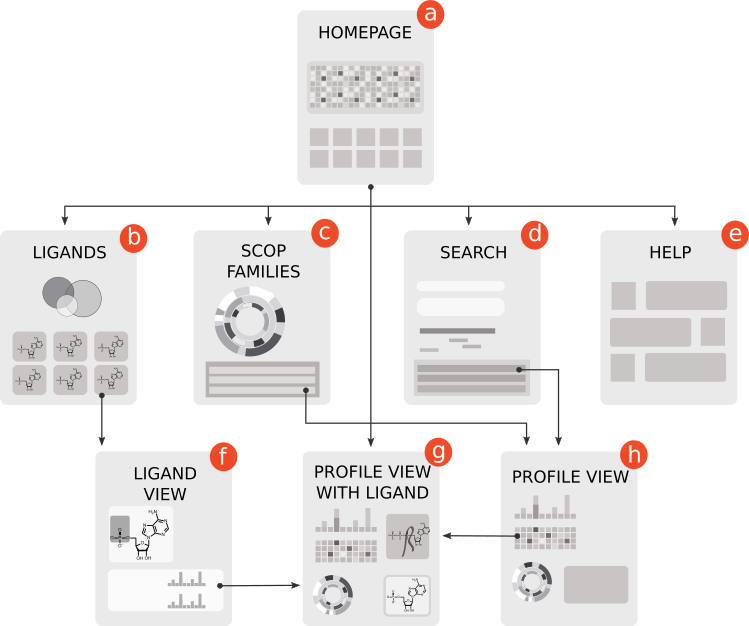
Ligands and cofactors
|
#
|
Full Name
|
Abbreviation
|
Molecular weight, g/mol
|
|
1
|
adenosine monophosphate
|
AMP
|
347.22
|
|
2
|
adenosine diphosphate
|
ADP
|
427.20
|
|
3
|
adenosine triphosphate
|
ATP
|
507.18
|
|
4
|
guanosine diphosphate
|
GDP
|
443.20
|
|
5
|
guanosine triphosphate
|
GTP
|
523.18
|
|
6
|
coenzyme A
|
CoA
|
767.53
|
|
7
|
acetyl coenzyme A
|
Acetyl-CoA
|
809.57
|
|
8
|
cyclic adenosine monophosphate
|
cAMP
|
329.05
|
|
9
|
cyclic guanosine monophosphate
|
cGMP
|
345.05
|
|
10
|
cyclic diadenosine monophosphate
|
c-di-AMP
|
692.47
|
|
11
|
cyclic diguanosine monophosphate
|
c-di-GMP
|
690.09
|
|
12
|
cytidine triphosphate
|
CTP
|
483.16
|
|
13
|
flavin adenine dinucleotide
|
FAD
|
785.55
|
|
14
|
flavin mononucleotide
|
FMN
|
456.34
|
|
15
|
coenzyme F420
|
F420
|
773.59
|
|
16
|
guanosine monophosphate
|
GMP
|
363.22
|
|
17
|
nicotinamide-adenine-dinucleotide
|
NAD
|
663.43
|
|
18
|
nicotinamide-adenine-dinucleotide phosphate
|
NADP
|
743.41
|
|
19
|
3'-phosphate-adenosine-5'-diphosphate
|
PAP
|
507.18
|
|
20
|
pyridoxal phosphate
|
PLP
|
247.14
|
|
21
|
3'-phosphate-adenosine-5'-phosphate sulfate
|
PPS
|
507.26
|
|
22
|
S-Adenosyl-L-methionine
|
SAM
|
398.44
|
|
23
|
thiamine diphosphate
|
TPP
|
425.31
|
|
24
|
alpha,beta-dihydroxyethyl-thiamin diphosphate
|
THDP
|
484.36
|
Example of protein with bound FMN ligand
We illustrate here how information collected in NB database can allow user to obtain a picture of nucleotide-containing ligand binding in different proteins. It shows how specific profiles find EFLs that interact with different parts of the ligand, delineating the key interactions between atoms of the ligands and amino acids of the protein.
Example 1.
Figure below shows a structure of dihydroorotate dehydrogenase (PDB ID: 2b4g) with six EFLs binding flavin mononucleotide (FMN). Sequences of these EFLs were found by corresponding profiles. In particular, profiles GAQTG, GGI, and GFGGGG found EFLs that interact with a phosphate moiety of FMN (blue, green, and orange loops, respectively). Profile MNAGCT – loop interacting with a sugar (red), profile SGKTRG – loop that interacts with a flavin (magenta), and profile PGKPP – loop that binds with both sugar and flavin (yellow).
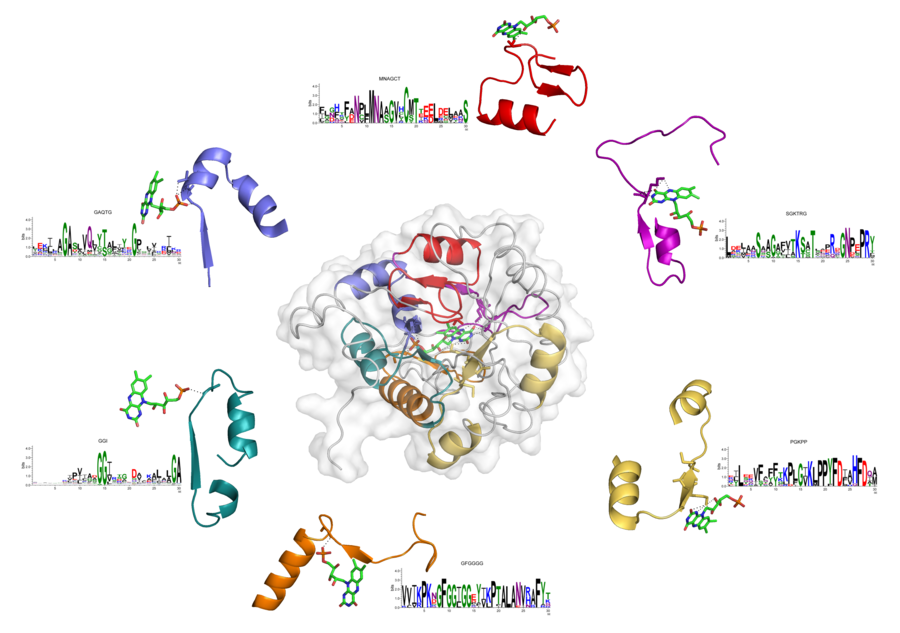
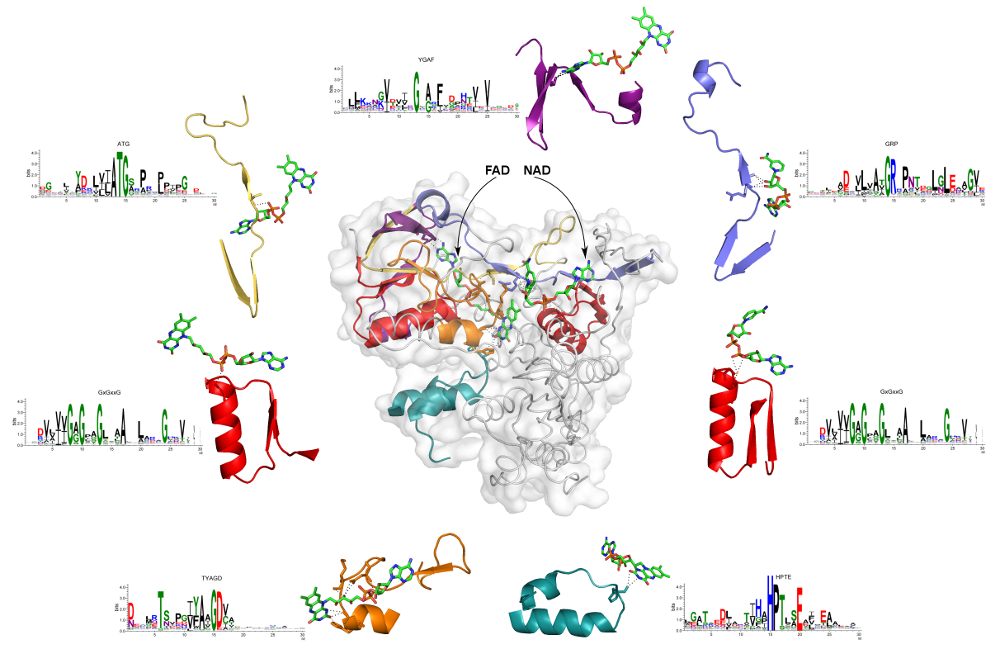
Example 2.
Figure below shows a structure of human dihydrolipoamide dehydrogenase (hE3, UniProt accession number is P09622, PDB ID is 1zmc) with two bound coenzymes: a non-covalently bound FAD and a transiently bound NAD+. This protein is an interesting example, because two different nucleotide-containing ligands are bound simultaneously. We detected seven EFLs interacting with these ligands: five loops provide binding of FAD, and two – NAD. There are two different EFLs binding the phosphate moieties in both FAD and NAD (red loops on the right and left, respectively). Remarkably, they are both representatives of the most common signature of the phosphate binding in dinucleotide-containing ligands – GxGxxG. Additionally, EFL (yellow) of the ATG profile interacts with the phosphate of FAD. Ribose of the NAD is bound to representative of the GRP profile (blue loop). Flavin is stabilized by interactions with loops matching to HPTE (cyan) and TYAGD (orange) profiles. The later also takes part in interactions with the sugar moiety. Profile YGAF detected the loop (purple) interacting with the base of FAD.
Important definitions
Elementary Function (EF)
is an elementary reaction or binding interaction that provides stabilization of the transition state [1] en route of the biochemical transformation.
Closed loop
or return of the polypeptide backbone with a typical size of 25–30 amino acid residues is a basic universal structural element of globular proteins originating from the polymer nature of polypeptide chains [2,3].
Elementary Functional Loop (EFL)
is defined as a structural-functional unit formed by the closed loop [2,3], carrying one or a few functional residues responsible for the corresponding elementary reaction or binding interactions [4,5]. EFL serves as a minimal functional building block in biochemical mechanisms.
Profile of the EFL
is a position specific scoring matrix (PSSM) representing an ensemble of multiple sequences of elementary functional loops [4].
Ligand parts and chemical moieties
We distinguish interactions with distinct chemical parts of nucleotide-containing ligands and cofactors, such as nitrogenous bases (adenine, guanine, cytosine), phosphate groups, ribose (or other sugars), and flavin, nicotinamide, thiamine, and other moieties. Ligand's atoms are named according to the PDB notation.
Database resources
PDB
the Protein Databank was used as a source of structural information on ligand-protein interactions. We identified hydrogen bonds with the ligand with the help of Chimera software. PDB also provides 2D views of ligand-protein interactions generated with PoseView software.
UniProt
is a comprehensive resource for sequence information. We used UniRef50 for deriving the EFL profiles. UniRef50 is a non-redundant collection of proteins with at most 50% pairwise sequence identity.
Download NBDB data in machine-readable formats
Annotation of molecular parts in ligands
Each ligand's atom is assigned a chemical part code. We distinguish the following parts: (B) base, (R) ribose, (P) phosphate, (F) flavin, (N) nicotinamide, (T) thiamine, (S) sulfur, (O) other.
PDB hits of EFL profiles
Tab-delimited file with columns: EFL profile name, PDB and chain, score, starting amino acid number of the match, match sequence, SCOP domain coordinates, SCOP superfamily code, SCOP family code, SCOP superfamily id, SCOP family id, SCOP superfamily name, SCOP family name.
All interactions between profiles and ligands
Each block of data starts with profile name and a list of PDB codes that were used as structural evidence. Location number is position within the profile. Each line of interaction is defined for molecular part (see the list of part codes above), atom name (according to PDB), and the number of interactions with the corresponding EFL profile position.
EFL profile PFM (example for profile ADEP)
Each position frequency matrix (PFM) is accessible in machine-readable format, where the rows are profile positions (counting from zero) and the columns are amino acids in alphabetical order. Each value represents frequency of an amino acid on the corresponding position.
In order to fetch the matrix files for other profiles change the URL accordingly.
References
- Jencks WP. Catalysis in chemistry and enzymology. Mineola NY, editor: Dover; 1987.
- Berezovsky IN, Grosberg AY, Trifonov EN. Closed loops of nearly standard size: common basic element of protein structure. FEBS letters 2000;466(2-3):283-286.
- Berezovsky IN, Trifonov EN. Van der Waals locks: loop-n-lock structure of globular proteins. Journal of molecular biology 2001;307(5):1419-1426.
- Goncearenco A, Berezovsky IN. Prototypes of elementary functional loops unravel evolutionary connections between protein functions. Bioinformatics 2010;26(18):i497-503.
- Goncearenco A, Berezovsky IN. Computational reconstruction of primordial prototypes of elementary functional loops in modern proteins. Bioinformatics 2011;27(17):2368-2375.